Systems, Synthetic, and Computational Bioengineering
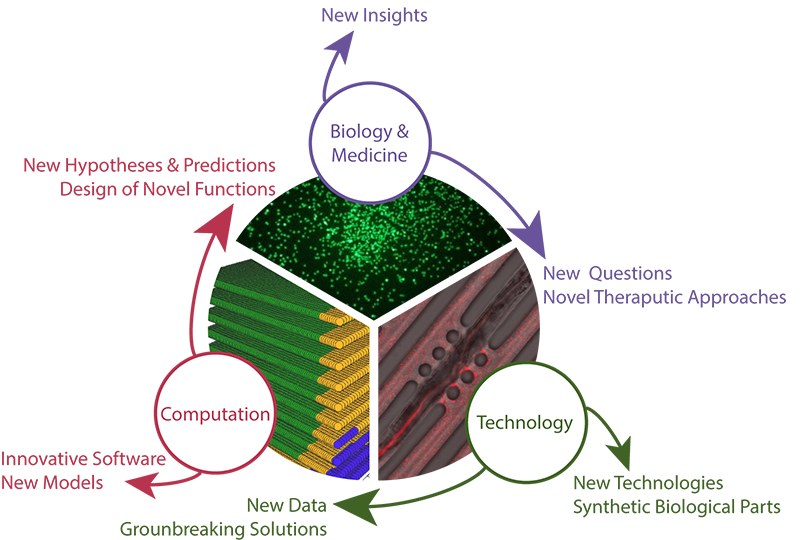
Research groups in systems, synthetic, and computational bioengineering apply engineering principles to model and understand complex biological systems, including differentiation and development, pathogenesis and cancer, and learning and behavior. This involves designing and implementing methods for procuring quantitative and sometimes very large data sets, as well as developing theoretical models and computational tools for interpreting these data.
Deciphering the workings of a biological system allows us to identify potential biomarkers and drug targets, to develop protocols for personalized medicine, and more. In addition, we use the design principles of biological systems we discover to engineer and refine new synthetic biological systems for clinical, agricultural, environmental, and energy applications.
Primary faculty contacts
 |
Assistant Research Professor Artificial Intelligent including Deep Learning and Machine Learning for Healthcare, Natural Language Processing (NLP) and Image Processing for Healthcare, Recommender Systems for Healthcare |
 |
Associate Professor Statistical and machine learning approaches to biological data; Gut-brain-microbe interactions; multi-cellular synthetic biology |
 |
University Distinguished Professor Theoretical approaches to the functional behavior of living systems. Cell signaling and cell motility. Cancer metastasis and the cancer-immune interaction. Drug resistance and epigenetics. |
 |
Assistant Professor Synthetic biology, protein engineering, microbiology, and cell signaling |
 |
Assistant Professor Computational systems biology, gene regulatory networks, single cell genomics |
|
|
Assistant Professor Computational prediction of molecular interactions at biological interfaces |
 |
Associate Professor Single-cell proteomics and analysis, cell signaling, systems biology |
 |
University Distinguished Professor Feedback control theory, systems biology, cancer, and biomedicine |
 |
Assistant Research Professor Systems biology of signaling networks in cancer (and) therapeutics, metastasis and embryonic development |
 |
Professor Computational modeling of the cardiac myocyte to understand the molecular basis of arrhythmias; machine learning in critical care medicine to identify those patients who require urgent care |
